Assessing Read Quality
Overview
Teaching: 30 min
Exercises: 20 minQuestions
How can I describe the quality of my data?
Objectives
Explain how a FASTQ file encodes per-base quality scores
Interpret a FastQC plot summarizing per-base quality across all reads
Use
forloops to automate operations on multiple files
Bioinformatics Workflows
When working with high-throughput sequencing data, the raw reads you get off of the sequencer will need to pass through a number of different tools in order to generate your final desired output. The execution of this set of tools in a specified order is commonly referred to as a workflow or a pipeline.
An example of the workflow we will be using for our variant calling analysis is provided below with a brief description of each step.
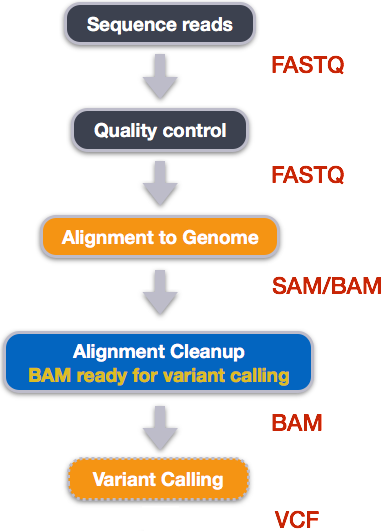
- Quality control – Assessing quality using FastQC
- Quality control – Trimming and/or filtering reads (if necessary)
- Align reads to reference genome
- Perform post-alignment clean-up
- Variant calling
These workflows in bioinformatics adopt a plug-and-play approach in that the output of one tool can be used as input to another tool without any extensive configuration. Having standards for data formats is what makes this feasible. Standards ensure that data is stored in a way that is generally accepted and agreed upon within the community. The tools that are used to analyze data at different stages of the workflow are therefore built under the assumption that the data will be provided in a specific format.
Quality Control
The first step in the variant calling workflow is to take the FASTQ files received from the sequencing facility and assess the quality of the sequence reads.
Details on the FASTQ format
Although it looks complicated (and it is), it’s easy to understand the FASTQ format with a little decoding. Some rules about the format include:
| Line | Description |
|---|---|
| 1 | Always begins with @ and then information about the read |
| 2 | The actual DNA sequence |
| 3 | Always begins with a + and sometimes the same info in line 1 |
| 4 | Has a string of characters which represent the quality scores; must have same number of characters as line 2 |
We can view the first complete read in one of the files our dataset by using head to
look at the first four lines.
$ head -n4 SRR098026.fastq
@SRR098026.1 HWUSI-EAS1599_1:2:1:0:968 length=35
NNNNNNNNNNNNNNNNCNNNNNNNNNNNNNNNNNN
+SRR098026.1 HWUSI-EAS1599_1:2:1:0:968 length=35
!!!!!!!!!!!!!!!!#!!!!!!!!!!!!!!!!!!
All but one of the nucleotides in this read are unknown (N). This is a pretty bad read!
Line 4 shows the quality for each nucleotide in the read. Quality is interpreted as the probability of an incorrect base call (e.g. 1 in 10) or, equivalently, the base call accuracy (e.g., 90%). To make it possible to line up each individual nucleotide with its quality score, the numerical score is converted into a code where each individual character represents the numerical quality score for an individual nucleotide. For example, in the line above, the quality score line is:
!!!!!!!!!!!!!!!!#!!!!!!!!!!!!!!!!!!
The # character and each of the ! characters represent the encoded quality for an
individual nucleotide. The numerical value assigned to each of these characters
depends on the sequencing platform that generated the reads. The sequencing machine
used to generate our data uses the standard Sanger quality PHRED score encoding, used
by Illumina version 1.8 onwards. Each character is assigned a quality score between 0
and 40 as shown in the chart below.
Quality encoding: !"#$%&'()*+,-./0123456789:;<=>?@ABCDEFGHI
| | | | |
Quality score: 0........10........20........30........40
Each quality score represents the probability that the corresponding nucleotide call is incorrect. This quality score is logarithmic, so a quality score of 10 reflects a base call accuracy of 90%, while a quality score of 20 reflects a base call accuracy of 99%. These probability values are the results from the base calling algorithm and dependent on how much signal was captured for the base incorporation.
Looking back at our read:
@SRR098026.1 HWUSI-EAS1599_1:2:1:0:968 length=35
NNNNNNNNNNNNNNNNCNNNNNNNNNNNNNNNNNN
+SRR098026.1 HWUSI-EAS1599_1:2:1:0:968 length=35
!!!!!!!!!!!!!!!!#!!!!!!!!!!!!!!!!!!
we can now see that the quality of each of the Ns is 0 and the quality of the only
nucleotide call (C) is also very poor (# = a quality score of 2). This is indeed a
very bad read.
Exercise
What is the last read in the
SRR098026.fastqfile? How confident are you in this read?Solution
$ tail -n4 SRR098026.fastq@SRR098026.249 HWUSI-EAS1599_1:2:1:2:1057 length=35 CNCTNTATGCGTACGGCAGTGANNNNNNNGGAGAT +SRR098026.249 HWUSI-EAS1599_1:2:1:2:1057 length=35 A!@B!BBB@ABAB#########!!!!!!!######The second half of this read is poor quality. Many of the positions are unknown (Ns) and the bases that we do have guesses for are of very poor quality (
#). However, the beginning of the read is fairly high quality. We will look at variations in position-based quality in just a moment.
Quality Encodings Vary
Although we’ve used a particular quality encoding system to demonstrate interpretation of read quality, different sequencing machines use different encoding systems. This means that, depending on which sequencer you use to generate your data, a
#may not be an indicator of a poor quality base call.This mainly relates to older Solexa/Illumina data, but it’s essential that you know which sequencing platform was used to generate your data, so that you can tell your quality control program which encoding to use. If you choose the wrong encoding, you run the risk of throwing away good reads or (even worse) not throwing away bad reads!
Assessing Quality using FastQC
In real life, you won’t be assessing the quality of your reads by visually inspecting your FASTQ files. Rather, you’ll be using a software program to assess read quality and filter out poor quality reads. We’ll first use a program called FastQC to visualize the quality of our reads. Later in our workflow, we’ll use another program to filter out poor quality reads.
FastQC has a number of features which can give you a quick impression of any problems your data may have, so you can take these issues into consideration before moving forward with your analyses. Rather than looking at quality scores for each individual read, FastQC looks at quality collectively across all reads within a sample. The image below shows a FastQC-generated plot that indicates a very high quality sample:
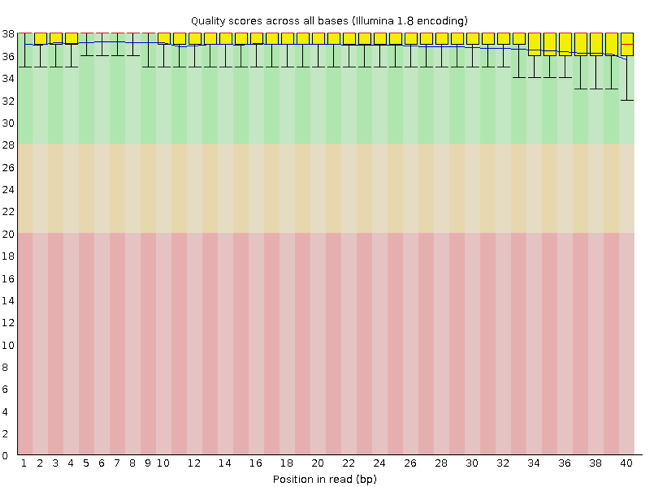
The x-axis displays the base position in the read, and the y-axis shows quality scores. In this example, the sample contains reads that are 40 bp long. For each position, there is a box-and-whisker plot showing the distribution of quality scores for all reads at that position. The horizontal red line indicates the median quality score and the yellow box shows the 2nd to 3rd quartile range. This means that 50% of reads have a quality score that falls within the range of the yellow box at that position. The whiskers show the range to the 1st and 4th quartile.
For each position in this sample, the quality values do not drop much lower than 32. This is a high quality score. The plot background is also color-coded to identify good (green), acceptable (yellow), and bad (red) quality scores.
Now let’s take a look at a quality plot on the other end of the spectrum.
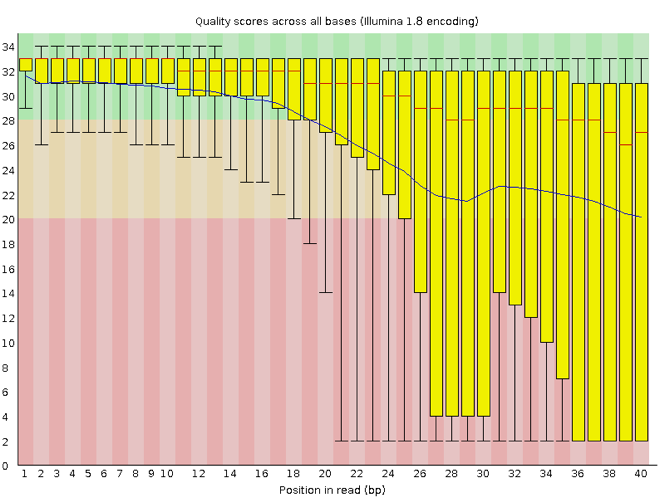
Here, we see positions within the read in which the boxes span a much wider range. Also, quality scores drop quite low into the “bad” range, particularly on the tail end of the reads. The FastQC tool produces several other diagnostic plots to assess sample quality, in addition to the one plotted above.
Running analyses on Crane
We are going to start doing analyses on our data. So far we’ve been doing all our work
on the Crane “login” nodes. It’s fine to use the nodes for simple tasks like examining
files with less, editing files with nano, moving files, but for analyses we need
to have the work done on the “compute” nodes. For this workshop we will be using the
interactive queue. With one command you will start an interactive
job, be logged into one of the compute nodes, mount the /common/ directory
that we will need later on, and set a timeout of our sesson at 3 hours.
The rest of our work will continue on the compute note.
$ srun --pty --time=3:00:00 --license=common $SHELL
[username@cNNNN]$
You will notice that the command prompt will change to say that you are now on a
compute node (@cNNN).
$ pwd
/work/group/username
We’ll make sure to cd back to our data/ directory below.
Another important thing to remember is to load any modules we need. We’ll do that before we run each analysis.
Running FastQC
We will be using the FASTQ dataset you transferred yesterday. Please navigate to the files.
$ cd /work/group/username/dc_workshop/data/untrimmed_fastq/
Your directory should look like:
$ ls -F
SRR097977.fastq* SRR098027.fastq* SRR098281.fastq*
SRR098026.fastq* SRR098028.fastq* SRR098283.fastq*
Exercise
How many FASTQ files are in this dataset? How big are the files? (Hint: Look at the options for the
lscommand to see how to show file sizes.)Solution
$ ls -l -h-rw-r--r-- 1 username group 840M Jul 30 2015 SRR097977.fastq -rw-r--r-- 1 username group 3.4G Jul 30 2015 SRR098026.fastq -rw-r--r-- 1 username group 875M Jul 30 2015 SRR098027.fastq -rw-r--r-- 1 username group 3.4G Jul 30 2015 SRR098028.fastq -rw-r--r-- 1 username group 4.0G Jul 30 2015 SRR098281.fastq -rw-r--r-- 1 username group 3.9G Jul 30 2015 SRR098283.fastqThere are six FASTQ files ranging from 840M (840MB) to 4.0G (4.0GB)
To run the FastQC program, we need to load the fastqc module (fastqc). FastQC
can accept multiple file names as input, so we can use the *.fastq wildcard to run
FastQC on all of the FASTQ files in this directory.
$ module load fastqc
$ fastqc *.fastq
You will see an automatically updating output message telling you the progress of the analysis. It will start like this:
Started analysis of SRR097977.fastq
Approx 5% complete for SRR097977.fastq
Approx 10% complete for SRR097977.fastq
Approx 15% complete for SRR097977.fastq
Approx 20% complete for SRR097977.fastq
Approx 25% complete for SRR097977.fastq
Approx 30% complete for SRR097977.fastq
Approx 35% complete for SRR097977.fastq
Approx 40% complete for SRR097977.fastq
Approx 45% complete for SRR097977.fastq
Approx 50% complete for SRR097977.fastq
...
In total, it should take about five minutes for FastQC to run on all six of our FASTQ files. When the analysis completes, your prompt will return. So your screen will look something like this:
Approx 80% complete for SRR098283.fastq
Approx 85% complete for SRR098283.fastq
Approx 90% complete for SRR098283.fastq
Approx 95% complete for SRR098283.fastq
Analysis complete for SRR098283.fastq
The FastQC program has created several new files within our
data/untrimmed_fastq/ directory.
$ ls
SRR097977.fastq SRR098027.fastq SRR098281.fastq
SRR097977_fastqc/ SRR098027_fastqc/ SRR098281_fastqc/
SRR097977_fastqc.zip SRR098027_fastqc.zip SRR098281_fastqc.zip
SRR098026.fastq SRR098028.fastq SRR098283.fastq
SRR098026_fastqc/ SRR098028_fastqc/ SRR098283_fastqc/
SRR098026_fastqc.zip SRR098028_fastqc.zip SRR098283_fastqc.zip
For each input FASTQ file, FastQC has created a .zip file and a
directory contain an .html report file. The .zip file extension
indicates that this is actually a compressed set of multiple output
files. We’ll be working with these output files soon.
The .html file is a stable webpage displaying the summary
report for each of our samples.
We want to keep our data files and our results files separate, so we
will move these output files into a new directory within our results/ directory.
$ mkdir /work/group/username/dc_workshop/results/fastqc_untrimmed_reads
$ mv *.zip /work/group/username/dc_workshop/results/fastqc_untrimmed_reads/
$ mv *fastqc/ /work/group/username/dc_workshop/results/fastqc_untrimmed_reads/
Now we can navigate into this results/ directory and do some closer
inspection of our output files.
$ cd /work/group/username/dc_workshop/results/fastqc_untrimmed_reads/
Viewing HTML files
If we were working on our local computers, we’d be able to display each of these HTML files as a webpage:
$ open SRR097977_fastqc/fastqc_report.html
However, if you try this on our Crane, you’ll get an error:
Couldn't get a file descriptor referring to the console
This is because Crane doesn’t have any web browsers installed on it, so the remote computer doesn’t know how to open the file. We want to look at the webpage summary reports, so let’s transfer them to our local computers (i.e. your laptop).
To transfer a file from a remote server to our own machines, we will
use a variant of the cp command called scp. The “s” stands for
“secure” – this is a secure version of copying (cp).
For the cp command, the syntax was:
$ cp my_file new_location
The syntax for scp is the same, but now my_file and
new_location are on separate computers, so we need to give an
absolute path, including the name of our remote computer. First we
will make a new directory on our computer to store the HTML files
we’re transferring. Let’s put it on our Desktop for now.
Now we can transfer our HTML files to our local computer using scp.
On Macs or Linux
Open a new tab in your terminal program (you can use the pull down menu at the top of your screen or the Ctrl+t keyboard shortcut) and type:
$ mkdir ~/Desktop/fastqc_html
$ scp username@crane.unl.edu:/work/group/username/dc_workshop/results/fastqc_untrimmed_reads/*.zip ~/Desktop/fastqc_html/
On Windows
Open a Windows command prompt window by typing Windows+r (the Windows key
and the ‘r’ key at the same time). You should see a “Run” window open. In the box
enter: cmd, a new cmd.exe window should open.
Create a new folder on your desktop:
$ mkdir %HOMEPATH%\Desktop\fastqc_html
(%HOMEPATH% is roughly the same as ~ in Unix based systems.)
Now we can transfer our HTML files to our local computer using pscp. (Note that Linux systems use the forward slash, /, for directories while Windows uses the back slash, \, check that you are using the right ones in the command below.)
> pscp username@crane.unl.edu:/work/group/username/dc_workshop/results/fastqc_untrimmed_reads/*.zip %HOMEPATH%\Desktop\fastqc_html\
Back to everyone
This looks really complicated, so let’s break it down. The first part
of the command username@crane.unl.edu is the address for your remote computer.
Make sure you replace username with your actual username.
The second part starts with a : and then gives the absolute path
of the files you want to transfer from your remote computer. Don’t
forget the :. We used a wildcard (*.zip) to indicate that we want all of
the zip files.
The third part of the command gives the absolute path of the location
you want to put the files. This is on your local computer and is the
directory we just created ~/Desktop/fastqc_html/.
You should see a status output like this:
SRR097977_fastqc.zip 100% 130KB 130.1KB/s 00:00
SRR098026_fastqc.zip 100% 183KB 182.8KB/s 00:00
SRR098027_fastqc.zip 100% 129KB 129.0KB/s 00:00
SRR098028_fastqc.zip 100% 192KB 191.7KB/s 00:00
SRR098281_fastqc.zip 100% 195KB 195.0KB/s 00:00
SRR098283_fastqc.zip 100% 192KB 192.0KB/s 00:00
Navigate to this fastqc_html/ directory in your usual file explorer and
unzip the files as you usually would. We will learn how to programmatically
unzip this files in just a little bit.
Viewing on Mac and Linux systems:
$ cd ~/Desktop/fastqc_html/
$ open */*.html
Viewing on Windows:
> cd %HOMEPATH%\Desktop\fastqc_html
> SRR097977_fastqc.html
> SRR098026_fastqc.html
> SRR098027_fastqc.html
> SRR098028_fastqc.html
> SRR098281_fastqc.html
> SRR098283_fastqc.html
Your computer will open each of the HTML files in your default web browser. Depending on your settings, this might be as six separate tabs in a single window or six separate browser windows.
Exercise
Discuss your results with a neighbor. Which sample(s) looks the best in terms of per base sequence quality? Which sample(s) look the worst?
Solution
SRR097977andSRR098027are the best. The other four samples are all pretty bad.
Unzipping Compressed Files
Now that we’ve looked at our HTML reports to get a feel for the data,
let’s look more closely at the other output files. Go back to the tab
in your terminal program that is connected to Crane and make sure you’re in
our results/ subdirectory.
$ cd /work/group/username/dc_workshop/results/fastqc_untrimmed_reads/
$ ls -F
SRR097977_fastqc/ SRR098026_fastqc.zip SRR098028_fastqc/
SRR098281_fastqc.zip SRR097977_fastqc.zip SRR098027_fastqc/
SRR098028_fastqc.zip SRR098283_fastqc/ SRR098026_fastqc/
SRR098027_fastqc.zip SRR098281_fastqc/ SRR098283_fastqc.zip
Our .zip files are compressed files. They each contain multiple
different types of output files for a single input FASTQ file. To
view the contents of a .zip file, we can use the program unzip
to list without decompressing these files.
Let’s try doing them all at once using a wildcard.
$ unzip -l *.zip
Archive: SRR097977_fastqc.zip
Length Date Time Name
--------- ---------- ----- ----
--------- -------
0 0 files
This didn’t work. What happened here is unzip -l expects one .zip argument and we
gave it six. We could go through and unzip each file one at a time, but this is very
time consuming and error-prone. Someday you may have 500 files to unzip!
A more efficient way is to use a for loop to iterate through all of
our .zip files. Let’s see what that looks like and then we’ll
discuss what we’re doing with each line of our loop.
$ for filename in *.zip
> do
> unzip -l $filename
> done
When the shell sees the keyword for, it knows to repeat a command (or group of
commands) once for each item in a list. Each time the loop runs (called an iteration),
an item in the list is assigned in sequence to the variable, and the commands
inside the loop are executed, before moving on to the next item in the list.
Inside the loop, we call for the variable’s value by putting $ in front of it.
The $ tells the shell interpreter to treat the variable as a variable name and
substitute its value in its place, rather than treat it as text or an external command.
In this example, the list is six filenames (one filename for each of our .zip files).
Each time the loop iterates, it will assign a file name to the variable filename and
run the unzip command. The first time through the loop, $filename is
SRR097977_fastqc.zip. The interpreter runs the command unzip on
SRR097977_fastqc.zip. For the second iteration, $filename becomes
SRR098026_fastqc.zip. This time, the shell runs unzip on SRR098026_fastqc.zip.
It then repeats this process for the four other .zip files in our directory.
Follow the Prompt
The shell prompt changes from
$to>and back again as we were typing in our loop. The second prompt,>, is different to remind us that we haven’t finished typing a complete command yet. A semicolon,;, can be used to separate two commands written on a single line.
Same Symbols, Different Meanings
Here we see
>being used a shell prompt, whereas>is also used to redirect output. Similarly,$is used as a shell prompt, but, as we saw earlier, it is also used to ask the shell to get the value of a variable.If the shell prints
>or$then it expects you to type something, and the symbol is a prompt.If you type
>or$yourself, it is an instruction from you that the shell to redirect output or get the value of a variable.
We have called the variable in this loop filename in order to make its purpose
clearer to human readers. The shell itself doesn’t care what the variable is called;
if we wrote this loop as:
$ for x in *.zip
> do
> unzip -l $x
> done
or:
$ for temperature in *.zip
> do
> unzip -l $temperature
> done
it would work exactly the same way. Don’t do this.
Programs are only useful if people can understand them,
so meaningless names (like x) or misleading names (like temperature)
increase the odds that the program won’t do what its readers think it does.
Multipart commands
The
forloop is interpreted as a multipart command. If you press the up arrow on your keyboard to recall the command, it will be shown like so:$ for filename in *.zip; do unzip -l $filename; doneWhen you check your history later, it will help your remember what you did!
When we run our for loop, you will see output that starts like this:
Archive: SRR097977_fastqc.zip
Length Date Time Name
--------- ---------- ----- ----
0 12-13-2018 13:28 SRR097977_fastqc/
0 12-13-2018 13:28 SRR097977_fastqc/Icons/
0 12-13-2018 13:28 SRR097977_fastqc/Images/
1561 12-13-2018 13:28 SRR097977_fastqc/Icons/error.png
1715 12-13-2018 13:28 SRR097977_fastqc/Icons/tick.png
1450 12-13-2018 13:28 SRR097977_fastqc/Icons/warning.png
1197 12-13-2018 13:28 SRR097977_fastqc/Icons/fastqc_icon.png
487 12-13-2018 13:28 SRR097977_fastqc/summary.txt
.
.
.
The unzip program is listing the content of the .zip files and if we ran unzip
without the -l flag would decompress these file, creating a new directory (with
subdirectories) for each of our samples, to store all of the different output that is
produced by FastQC. There are a lot of files here. The one we’re going to focus on is
the summary.txt file.
Understanding FastQC Output
If you list the files in our directory now using ls -F you will see:
SRR097977_fastqc/ SRR098026_fastqc.zip SRR098028_fastqc/ SRR098281_fastqc.zip
SRR097977_fastqc.zip SRR098027_fastqc/ SRR098028_fastqc.zip SRR098283_fastqc/
SRR098026_fastqc/ SRR098027_fastqc.zip SRR098281_fastqc/ SRR098283_fastqc.zip
Let’s see what files are present within one of these output directories.
$ ls -F SRR097977_fastqc/
fastqc_data.txt fastqc_report.html Icons/ Images/ summary.txt
Use less to preview the summary.txt file for this sample.
$ less SRR097977_fastqc/summary.txt
PASS Basic Statistics SRR097977.fastq
WARN Per base sequence quality SRR097977.fastq
PASS Per sequence quality scores SRR097977.fastq
PASS Per base sequence content SRR097977.fastq
PASS Per base GC content SRR097977.fastq
PASS Per sequence GC content SRR097977.fastq
PASS Per base N content SRR097977.fastq
PASS Sequence Length Distribution SRR097977.fastq
WARN Sequence Duplication Levels SRR097977.fastq
WARN Overrepresented sequences SRR097977.fastq
FAIL Kmer Content SRR097977.fastq
The summary file gives us a list of tests that FastQC ran, and tells
us whether this sample passed, failed, or is borderline (WARN).
Documenting Our Work
We can make a record of the results we obtained for all our samples
by concatenating all of our summary.txt files into a single file
using the cat command. We’ll call this fastqc_summaries.txt and move
it to /work/group/username/dc_workshop/docs.
$ cat */summary.txt > /work/group/username/dc_workshop/docs/fastqc_summaries.txt
Exercise
Which samples failed at least one of FastQC’s quality tests? What test(s) did those samples fail?
Solution
We can get the list of all failed tests using
grep.$ cd /work/group/username/dc_workshop/docs $ grep FAIL fastqc_summaries.txtFAIL Per tile sequence quality SRR097977.fastq FAIL Per tile sequence quality SRR098026.fastq FAIL Overrepresented sequences SRR098026.fastq FAIL Kmer Content SRR098026.fastq FAIL Per base sequence quality SRR098027.fastq FAIL Per tile sequence quality SRR098027.fastq FAIL Kmer Content SRR098027.fastq FAIL Per tile sequence quality SRR098028.fastq FAIL Overrepresented sequences SRR098028.fastq FAIL Kmer Content SRR098028.fastq FAIL Overrepresented sequences SRR098281.fastq FAIL Kmer Content SRR098281.fastq FAIL Overrepresented sequences SRR098283.fastq FAIL Kmer Content SRR098283.fastqIf we want to see all the files that failed at least one test, we can use a combination of
grep,cut,sortanduniq.$ grep FAIL fastqc_summaries.txt | cut -f 3 | sort | uniqSRR097977.fastq SRR098026.fastq SRR098027.fastq SRR098028.fastq SRR098281.fastq SRR098283.fastqAll of our samples failed at least one test. If we want to see a table showing which tests failed, we can use the same command we used above, but this time extract the second field with
cut(instead of the third) and add the-coption touniqto count the number of times each unique value appears.$ grep FAIL fastqc_summaries.txt | cut -f 2 | sort | uniq -c6 Kmer Content 4 Overrepresented sequences 1 Per base sequence quality 4 Sequence Duplication Levels
Key Points
Quality encodings vary across sequencing platforms
forloops let you perform the same set of operations on multiple files with a single command